
Figure 1: Suyama's DNA computer
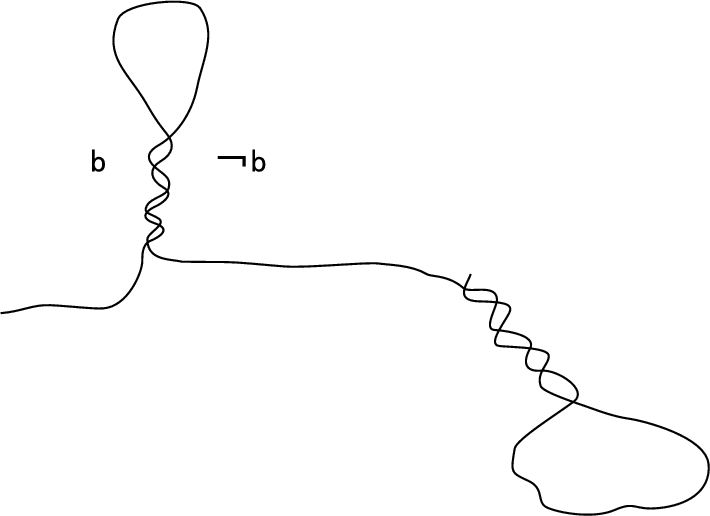
Figure 2: Molecule representing a logical formula with conflicting assignment b and !b (not b).
(Japanese page is here.)
Professor, Graduate School of Science, The University of Tokyo
Masami Hagiya
http://hagi.is.s.u-tokyo.ac.jp/MCP/
Molecular Computation Project (MCP) is an attempt to harness the computational power of molecules for information processing. In other words, it is a trial to develop a general-purpose computer with molecules [hagiya01keisokutoseigyo ][hagiya00shingakukaishi ] [hagiya00identekialgorithm ][Hagiya99NGC]. The idea of computing with molecules had not been truly realized until 1994, when L. Adleman published a breakthrough for making a general-purpose computer with biological molecules (DNA) [Adleman94Science]. Since then, the word `DNA computation' became widespread for the meaning of computation with DNA molecules.
Information processing on the molecular scale has been sought in several ways other than Adleman's, but the DNA computation is inherently different from other previous approaches: it aims the construction of a general-purpose computer based on the theory of universal computation. This goal seems to be hinted by the nature of DNA molecules, that is, an arbitrary concatenation of four natural bases forms one DNA sequence. We call this character of DNA as `combinatorial complexity'. Because of this complexity, DNA sequences can hold information of arbitral complexity by freely chaining four natural bases. Similarly, biological molecules such as RNA and proteins are appropriate for molecular computation, because they share this combinatorial complexity. It is worth mentioning that the combinatorial complexity underlies the complexity of life.
Molecular computation is a part of the larger body, `computational life science'. The goal of computational life science is to understand life systems from the perspective of theory of computation, and to apply its research result to bioengineering. In the real world, there exists a hierarchy of life systems. Its simplistic classification is molecules, organelle, cells, tissues (especially brain), individuals, societies, and ecosystems. In computational life science, each level in this hierarchy is the subject of computational investigation and is the target of engineering. Here, the computational investigation covers models of computation, and the analysis of their computational power or efficiency. Likewise, the engineering covers the construction of artificial systems as a research application. From this perspective, molecular computation shapes the most fundamental research embodying the hierarchy of life systems.
Molecular computation particularly focuses on the computational power of molecules, especially that of biological molecules, and attempts to realize information processing which maximally exploits the computational power of molecules. By using molecules, we expect to materialize faster (massively parallel), smaller (nano-scale), and cost efficient (energy-saving) information processing. Moreover, we also expect the emergence of brand-new technology of information processing, in particular, a new model of computation. The molecular implementation of a new information processing system is one major aim of molecular computation.
On the other hand, traditional semiconductor technology also foresees the construction of logic gates on the scale of molecules. This technology is called `molecular electronics', and a computer based on the molecular electronics is also called a molecular computer [Reed00nikkeiscience]. The differences between molecular computation and molecular electronics are as follows.
In 1996, Japan Society for the Promotion of Science (JSPS) started Research for the Future (RFTF) Program. Among its project fields, `Information on Life Systems' is responsible for the development of computational life science in Japan. The field is led by Yuichiro Anzai, the committee chairman and the dean of science and technology faculty at Keio university. The field consists of several research projects working on computational life science from various perspectives; our project, `Theory and Construction of Molecular Computers', directly targets molecular computation, and the following projects pursue related topics. `Biologically Inspired Adaptive Systems' (led by Prof. Kobayashi) explores a wide range of evolutionary computation and is firmly connected with our project through several ongoing joint researches. `Development of Artificial Cellular Device' (led by Prof. Ikuta) develops so-called "chemical IC", whose technology is indispensable for the high-performance and large-scale molecular computation. `Design and Fabrication of an Interface System for a Life Information Processor and External Environment' (led by Prof. Yamakawa and Prof. Haga) focuses on the signal transduction, especially its membrane receptors, in its biochemical approach. The membrane proteins are expected to function as I/O devices of a future cellular computer, where signal transduction functions as computation in a living cell.
The "Theory and Construction of Molecular Computers" project started in 1996 and ended in March 2001. Its members were:
Masami Hagiya (Computer Science, Graduate School of Information Science and Technology, University of Tokyo)
Takashi Yokomori (Computer Science, Department of Mathematics, School of Education, Waseda University)
Satoshi Kobayashi (Computer Science, Department of Computer Science, Faculty of Electro-Communications, University of Electro-Communications)
Yasubumi Sakakibara (Computer Science, Department of Information Sciences, Tokyo Denki University)
Akira Suyama (Biophysics, Graduate School of Art and Sciences, University of Tokyo)
Kensaku Sakamoto (Biochemistry, Graduate School of Science, University of Tokyo)
Yuzuru Fushimi (Biophysics, Faculty of Technology, Saitama University)
Masayuki Yamamura (Systems Science, Interdisciplinary Graduate School of Science and Engineering, Tokyo Institute of Techonology)
Masanori Arita (Genome Informatics, Computational Biology Research Center, Advanced Institute of Science and Technology)
Akio Nishikawa (Computer Science, Department of Electronics and Information Technology, Osaka Electro-Communication Junior College)
John Rose (Computer Science)
Our project is notable for its successful cooperation between researchers in information science (computer science and artificial intelligence) and those in biology (biophysics and biochemistry).
Before introducing main achievements of our project, this section briefly summarizes the global trend of molecular computation.
Arguably, DNA computation, or computation with DNA molecules, originates in a single paper in Science journal submitted by L. Adleman in 1994 [Adleman94Science]. He proposed a molecular algorithm to solve Hamiltonian Path Problem with DNA, and actually solved an instance of a directed graph with seven nodes. The idea of computing with molecules was not new by that time, but Adleman's approach outperformed previous ideas in that he exploited the massive parallelism of DNA molecules and that he actually demonstrated the algorithm by doing biological experiments by himself.
Adleman's work thus inseminated the field of DNA computation. Since 1995, the workshop `International Meeting on DNA Based Computers' started annually with the support of DIMACS (Center for Discrete Mathematics and Theoretical Computer Science), where more than 100 researchers participate in animated discussions every year. In addition, special sessions on DNA computation appeared in international conferences for genome informatics (including Pacific Symposium on Biocomputing) and for evolutionary computation (including Genetic and Evolutionary Computation Conference). Many achievements have been published in high-level academic journals including Nature, Science, Nucleic Acids Research, Journal of Computational Biology, Journal of Combinatorial Optimization, BioSystems, IEEE Transactions on Evolutionary Computation, and Soft Computing. Several patented technologies are also born from DNA computation.
Adleman's work is noteworthy as the starting point of this research field, for which he introduced one clear goal: faster solution of NP-complete problems. However, his argument overly emphasized the massive parallelism of molecules. As a result, many people were misled to the idea that solving combinatorial problems, including Hamiltonian Path Problem, faster than traditional digital computers is the only goal of molecular computation. Since the current technology is far behind this goal, the doubtful impression of molecular computation spread out.
Coherent understanding on the computational power of molecules can be applicable, however, to a broader research field related with biological molecules. For example, the following DARPA-NSF consortium carried out a wide range of attempts, in addition to using the massive parallelism, to exploit the computational power of molecules.
In the United States, the consortium for "Biomolecular Computation" started under DARPA and NSF, and carried out various attempts to grip the computational power of molecules. Led by J. Reif at Duke University, the consortium started in 1997 and ended in September 2000 [Reif99DARPA].
The code-name of the consortium is ``Prototyping Biomolecular Computations''. It tried to exploit not only the high-performance computation with the massive parallelism of molecules, but also the energy-saving computation using reactions on nano-scale.
Among several application areas of molecular computation, the technology of `nano-fabrication' is becoming an active research area. It is considered a part of nano-technology. Since DNA is the popular molecular tool, people call it `DNA nano-technology'. Its research result is applied not only to nano-fabrication but to nano-machine, i.e. the study on mechanistic machines on nano-scale.
Another promising application area of molecular computation is the genomic analysis such as DNA fingerprinting. In the consortium, a simple sensing technique such as DNA chip is enhanced with more intelligent measuring technology using molecular computation. Such biotechnical application of molecular computation is now called `computationally inspired biotechnology'.
In Europe, G. Rozenberg at Leiden University promotes a consortium on molecular computation. It headquarters at Center for Natural Computing in Leiden, and many research groups in Europe participate.
The research groups in Europe characteristically emphasize the theoretical aspects of molecular computation. In particular, Turing computability and computational complexity of molecular reactions are actively studied in relation to formal language theory, and a number of new results have been reported.
This section summarizes the main research results of our project.


>From its start, our project has been actively communicating with research groups abroad. Several joint research are ongoing, including the one for molecular memory, the cooperative work of Yamamura and Head (DARPA-NSF consortium). We also invited several researchers from the DARPA-NSF consortium in the United States and from the research community in Europe. Those invited include J. Reif at Duke University, the leader of the DARPA-NSF consortium in the United States, E. Winfree at California Institute of Technology, a young genius whose leading role in the DARPA consortium is prominent, and G. Paun, a masterly figure of formal language theory on DNA.
It is no longer believed that DNA computers will solve NP-complete problems faster than traditional digital computers; modern computers can solve the satisfiability problem of more than several hundred variables without errors. To match their speed, DNA computers would undergo incredible amount of breakthrough for their algorithms and for their implementation.
Researchers are now acknowledging that it is a bad idea to make molecular computers compete with digital computers on the same problem domain. According to their opinion, it is better to regard NP-complete problems as mere benchmarks to evaluate molecular computers.
Thus, it is an outdated idea to compare molecular computers with digital computers. Molecular computation should grow into a comprehensive study, from basics to applications, aiming at the information processing on molecular scale. Applications to biotechnology and nano-technology have been already started. In particular, the application to biotechnology is about to be realized, mainly because molecular computation uses biological molecules.
For example, Suyama's group tries to use their DNA computer for the analysis with DNA chips. The DNA chip of Suyama's group is called `universal chip', which is designed not to directly measure raw genetic information from cells, but to indirectly measure designed sequences, DNA Coded Number, translated from the raw information. [Suyama00CCMB]. DNA Coded Number is designed with techniques in DNA computation so that their interaction to one another is minimal, and that their amplification rate in PCR is uniform. Suyama and Sakakibara further proposed "intelligent DNA chip", which can perform logical reasoning and learning by using DNA computation on DNA Coded Number. [Sakakibara00GIW]. This is a typical application of molecular computation to biotechnology. Recently, biotechnology using molecular computation is called computationally inspired biotechnology.
Nano-technology, including molecular electronics, is an important application area too. DNA tile by E. Winfree is one such nano-technology with DNA (DNA nano-technology). A DNA tile can contain variable sequences at its single-stranded terminals, thus possessing combinatorial complexity. These tiles can self-assemble not only to a regular pattern but to a structure designated by a specific algorithm implemented in their single-stranded terminals. Self-assembly of this type is called algorithmic assembly. Algorithmic assembly can be used to design a template for placing molecular logic gates in molecular electronics.
Finally, let us mention one dreamlike perspective: medical application. Molecular computation has been studying an autonomous computation, such as whiplash PCR, which can change its state according to its environment. If we can realize a molecular machine which can measure its environmental factors and process information accordingly, then such a cellular machine opens a way to the medical application. Perhaps an elaborate E.coli engineered with molecular computation would diagnose our body by processing information on molecular scale, and would synthesize and exude appropriate medicines autonomously.